Random Walks in Higher Dimensions
So I was thinking about the dynamics of training, and looking for a way to model some phenomenon I had observed.
Initially, I had dismissed Brownian motion/random walk as an option because the trend was clearly linear — not square root. But then I was asked what Brownian motion looked like in higher dimensions and I panicked.
I had assumed that Brownian motion was universal — that dimensionality didn’t enter into the equation (for average distance from the origin as a function of time). But now I was doubting myself.
So I did a quick derivation, which I’m sharing because it gave me a new way to interpret the chi-squared distribution. Here’s the set-up: we have a walker on a $D$-dimensional (square) lattice. At every timestep, the walker chooses uniformly among the $2D$ available directions to take one step.
This means that every dimension is sampled on average once every $D$ timesteps, so we can treat a $D$-dimensional walker after $N$ timesteps as a collection of $D$ independent $1$-dimensional walkers after $N/D$ timesteps.
If the $1$-dimensional walker’s probability distribution is just a Gaussian centered at $0$ with standard deviation $\sqrt N$, then the probability for the $D$-dimensional walker is the product of $D$ univariate gaussians centered at $0$ with standard deviation $\sqrt{{N}/{D}}$.
But that means that the expected distance traveled by the $D$-dimensional walker, $\sqrt{\mathbb E[|\vec X|^2]}$, is just the square root of the chi-squared distribution,
$$
Y=\sqrt {\sum_{i=1}^{D}X_{i}^{2}},
$$
scaled by a factor $\sqrt{N/D}$. (This is not the chi distribution, which we’d want if we were taking the square root before the expectation.)
In this context, $D$ is the number of degrees of freedom. After multiplying the mean of the chi distribution by our factor $\sqrt{N/D}$, we get
$$
\sqrt{\mathbb E[|\vec X|^{2}]}= \sqrt{D} \sqrt{\frac{N}{D}} = \sqrt{N}.
$$
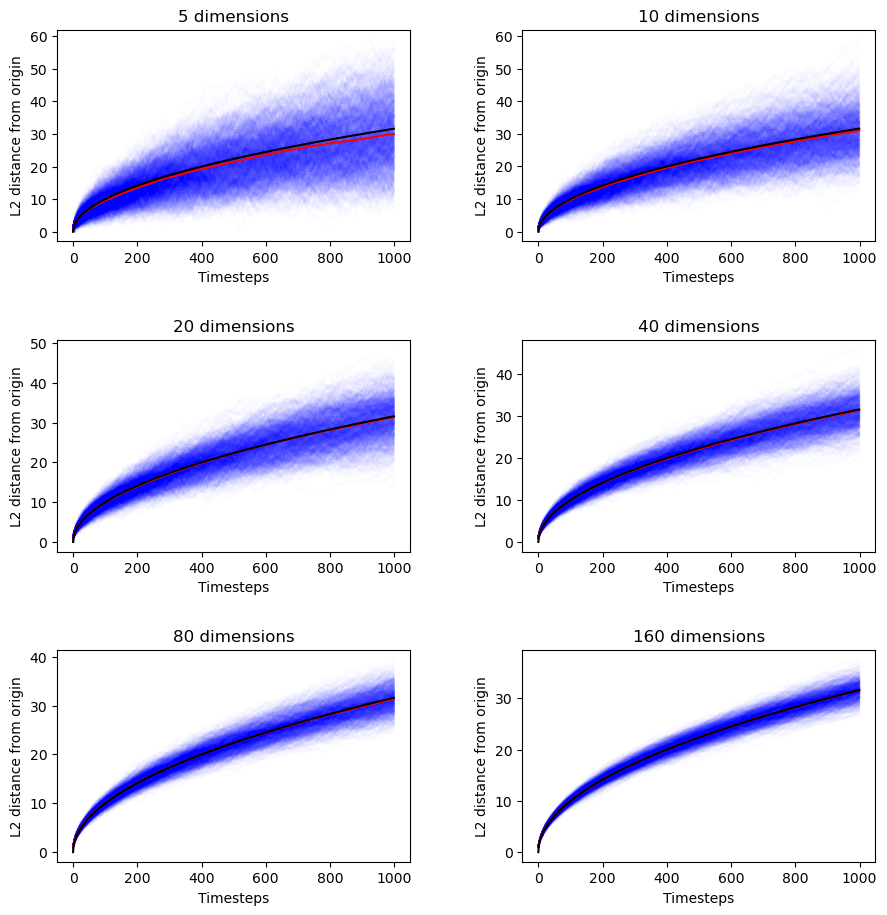
It’s independent of lattice dimensionality. Just as I originally thought before starting to doubt myself before going down this rabbit hole before reestablishing my confidence.
If that’s not enough for you, here are the empirical results.
def random_hyperwalk(n_dims: int, n_samples: int = 1_000, n_timesteps: int = 10_000) -> np.ndarray: state = np.zeros((n_samples, n_dims)) distances = np.zeros((n_samples, n_timesteps))
for t in tqdm(range(n_timesteps)): step = np.random.choice(np.arange(n_dims), n_samples) # print(step) directions = np.random.choice([-1, 1], n_samples) # print(directions)
# TODO: vectorize this for i in range(n_samples): state[i, step[i]] += directions[i]
# print(step, directions, state[:, step]) distances[:, t] = np.linalg.norm(state, axis=1)
return distances
def random_hyperwalk_scaling(n_dim_range: List[int], n_samples: int = 1_000, n_timesteps: int = 10_000): timesteps = np.arange(n_timesteps) predictions = np.sqrt(timesteps) trajectories = np.zeros((len(n_dim_range), n_samples, n_timesteps))
fig, ax = plt.subplots(len(n_dim_range) // 2, 2, figsize=(10, 10)) fig.tight_layout(pad=5.0)
for I, n_dims in enumerate(n_dim_range): i, j = I // 2, I % 2
trajectories[I, :, :] = random_hyperwalk(n_dims, n_samples, n_timesteps) mean_distances = np.mean(trajectories[I, :, :], axis=0)
for k in range(n_samples): ax[i][j].plot(timesteps, trajectories[I, k, :], color="blue", alpha=0.01)
ax[i][j].set_title(f"{n_dims} dimensions") ax[i][j].plot(timesteps, mean_distances, color="red") ax[i][j].plot(timesteps, predictions, color="black") ax[i][j].set_xlabel("Timesteps") ax[i][j].set_ylabel("L2 distance from origin")
plt.show()
random_hyperwalk_scaling([5, 10, 20, 40, 80, 160], n_samples=1000, n_timesteps=1000)